(Dr. Aldo Badano, Food and Drug Administration; affiliate faculty at the Fischell Department of Bioengineering)
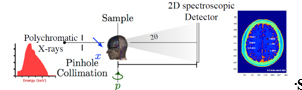 The presence of amyloid beta plaques in the human brain is considered one of the hallmark features of Alzheimer's disease (AD) and a promising biomarker for early diagnosis. In early AD stages, Aβ plaques are formed when Aβ peptides aggregate and accumulate in the brain to form higher-order fibrils with β-sheet structures. A structurally sensitive and high-resolution imaging method is required to detect plaques in the human brain and allow timely therapeutic interventions to prevent AD progression. To this end, we are designing a novel amyloid imaging system prototype based on small-angle x-ray scattering (SAXS) to quantify amyloid load without contrast agents based on the nanometer-scale structural features of plaques. REU student will have an opportunity to prepare human brain phantoms with amyloid plaque models and assist in testing the feasibility of detecting amyloid plaques using a newly designed amyloid imaging spectral SAXS prototype.
The presence of amyloid beta plaques in the human brain is considered one of the hallmark features of Alzheimer's disease (AD) and a promising biomarker for early diagnosis. In early AD stages, Aβ plaques are formed when Aβ peptides aggregate and accumulate in the brain to form higher-order fibrils with β-sheet structures. A structurally sensitive and high-resolution imaging method is required to detect plaques in the human brain and allow timely therapeutic interventions to prevent AD progression. To this end, we are designing a novel amyloid imaging system prototype based on small-angle x-ray scattering (SAXS) to quantify amyloid load without contrast agents based on the nanometer-scale structural features of plaques. REU student will have an opportunity to prepare human brain phantoms with amyloid plaque models and assist in testing the feasibility of detecting amyloid plaques using a newly designed amyloid imaging spectral SAXS prototype.
 The human gastrointestinal (GI) microbiome, often viewed as a complex organ itself, influences homeostasis and is implicated in many human diseases. There are few methodologies that enable resolution of its signaling, cell growth, and chemical environments even at the macro length scale. In the last several years however, researchers have developed micro- and meso-systems for interrogating GI tract biology. At the microscale, organ- or animal-on-a-chip methodologies have been developed for studying the GI tract. They provide first-of-their-kind access to biological function in user-controlled conditions. However, current approaches lack the ability to interrogate and modulate molecular signaling at cellular length scales and in real time. We have developed methodologies to combine synthetic biology with biofabrication to create smart hydrogels for interrogating molecular space. We do this by embedding “rewired” bacteria to serve as information translators into electrodeposited hydrogels that are assembled onto the sidewalls of a microfluidic channel (see Fig. 4). As a demonstration, we have assembled engineered E. coli to express blue fluorescent protein in response to a signal molecule, in this case the gratuitous lac operon inducer IPTG (Fig. 4c). In Fig. 4d, we show that one can layer cells using electrodeposition in configurations that enable visual access as well as chemical access so that cells can respond to molecular cues. In this REU project, using the same electrodeposition methodologies we have already developed in large systems, REU students will test the hypothesis that biologically active bacterial signaling molecules can be detected by a hydrogel bilayer that contains (i) signal sensing bacteria and (2) a catechol-modified chitosan layer incorporated onto the side wall of the channels shown in Fig. 4a,b. The students will assess whether the assembled cells can convert signal molecules to redox mediators in a manner that enables electrochemical detection. A successful result would be the first case of a biofabricated glucose sensor in an animal on a chip device. Such tests will be the first examples of the electrochemical interrogation of interkingdom biological signaling in an in vitro system. REU participants would conduct the majority of their work at the University of Maryland.
The human gastrointestinal (GI) microbiome, often viewed as a complex organ itself, influences homeostasis and is implicated in many human diseases. There are few methodologies that enable resolution of its signaling, cell growth, and chemical environments even at the macro length scale. In the last several years however, researchers have developed micro- and meso-systems for interrogating GI tract biology. At the microscale, organ- or animal-on-a-chip methodologies have been developed for studying the GI tract. They provide first-of-their-kind access to biological function in user-controlled conditions. However, current approaches lack the ability to interrogate and modulate molecular signaling at cellular length scales and in real time. We have developed methodologies to combine synthetic biology with biofabrication to create smart hydrogels for interrogating molecular space. We do this by embedding “rewired” bacteria to serve as information translators into electrodeposited hydrogels that are assembled onto the sidewalls of a microfluidic channel (see Fig. 4). As a demonstration, we have assembled engineered E. coli to express blue fluorescent protein in response to a signal molecule, in this case the gratuitous lac operon inducer IPTG (Fig. 4c). In Fig. 4d, we show that one can layer cells using electrodeposition in configurations that enable visual access as well as chemical access so that cells can respond to molecular cues. In this REU project, using the same electrodeposition methodologies we have already developed in large systems, REU students will test the hypothesis that biologically active bacterial signaling molecules can be detected by a hydrogel bilayer that contains (i) signal sensing bacteria and (2) a catechol-modified chitosan layer incorporated onto the side wall of the channels shown in Fig. 4a,b. The students will assess whether the assembled cells can convert signal molecules to redox mediators in a manner that enables electrochemical detection. A successful result would be the first case of a biofabricated glucose sensor in an animal on a chip device. Such tests will be the first examples of the electrochemical interrogation of interkingdom biological signaling in an in vitro system. REU participants would conduct the majority of their work at the University of Maryland.
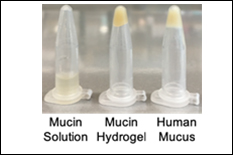 The constant generation and then movement of mucus from the lung airways to the throat, where it is then swallowed, is a key method by which the body protects against infection. However, in the airways of individuals with chronic lung diseases (e.g. asthma, chronic bronchitis, cystic fibrosis), build-up of mucus with abnormal biochemical and biophysical properties leads to airway obstruction and contributes to impaired lung function. Due to the natural heterogeneity of mucus derived from patients and animal models, we currently have a limited understanding of how mucus gel components impact the functional properties of mucus such as viscoelasticity for optimal mucus clearance and network architecture for pathogen/particulate capture. Toward this end, the goal of our NSF REU projects is to better understand the biophysical and biomolecular features of mucins that control their assembly into mucus. In our first REU student project, a bioengineered mucin-based hydrogel will be developed to mimic the physiological conditions under which the mucus gel network is assembled in health and disease. In a second project, an REU student will use CRISPR/Cas9 genetic engineering in order to design a tissue culture system with controlled airway mucus composition and function resembling that of a healthy and diseased airway. Together, this work will provide state-of-the art models that enable optimization and design of new therapies for chronic lung diseases. REU students engaged in this project will gain first-hand experience in biomaterial design, tissue culture, and/or genetic engineering. REU participants would conduct the majority of their work at the University of Maryland.
The constant generation and then movement of mucus from the lung airways to the throat, where it is then swallowed, is a key method by which the body protects against infection. However, in the airways of individuals with chronic lung diseases (e.g. asthma, chronic bronchitis, cystic fibrosis), build-up of mucus with abnormal biochemical and biophysical properties leads to airway obstruction and contributes to impaired lung function. Due to the natural heterogeneity of mucus derived from patients and animal models, we currently have a limited understanding of how mucus gel components impact the functional properties of mucus such as viscoelasticity for optimal mucus clearance and network architecture for pathogen/particulate capture. Toward this end, the goal of our NSF REU projects is to better understand the biophysical and biomolecular features of mucins that control their assembly into mucus. In our first REU student project, a bioengineered mucin-based hydrogel will be developed to mimic the physiological conditions under which the mucus gel network is assembled in health and disease. In a second project, an REU student will use CRISPR/Cas9 genetic engineering in order to design a tissue culture system with controlled airway mucus composition and function resembling that of a healthy and diseased airway. Together, this work will provide state-of-the art models that enable optimization and design of new therapies for chronic lung diseases. REU students engaged in this project will gain first-hand experience in biomaterial design, tissue culture, and/or genetic engineering. REU participants would conduct the majority of their work at the University of Maryland.
 Exosomes, a subset of cell-derived extracellular vesicles (EVs), have emerged as potential therapeutic vectors for a wide variety of applications. As alternatives to synthetic nanoparticles and cell therapies, exosomes have many potential benefits. Among these is the ability to transfer functional nucleic acids to recipient cells, as exosomes are physiological vectors for intercellular communication via nucleic acid transfer. However, exosomes also have shortcomings, especially their low potency due to containing relatively low amounts of nucleic acid cargo, especially non-coding RNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), which are critical to defining exosome bioactivity. Overcoming this limitation is crucial to enabling the therapeutic potential of exosomes. To this end, Dr. Jay and his group have studied how the biophysical culture microenvironment of exosome-producing cells can be manipulated to control non-coding RNA cargo and bioactivity of exosomes. This investigation is potentially transformative in identifying critical parameters that could be controlled towards development of rationally designed, scalable biomanufacturing approaches for therapeutic exosomes. REU students will have the opportunity to employ both 3D printing and microsystems design to test specific hypotheses related to the roles of biophysical parameters on exosome production and bioactivity. REU participants would conduct the majority of their work at the University of Maryland.
Exosomes, a subset of cell-derived extracellular vesicles (EVs), have emerged as potential therapeutic vectors for a wide variety of applications. As alternatives to synthetic nanoparticles and cell therapies, exosomes have many potential benefits. Among these is the ability to transfer functional nucleic acids to recipient cells, as exosomes are physiological vectors for intercellular communication via nucleic acid transfer. However, exosomes also have shortcomings, especially their low potency due to containing relatively low amounts of nucleic acid cargo, especially non-coding RNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), which are critical to defining exosome bioactivity. Overcoming this limitation is crucial to enabling the therapeutic potential of exosomes. To this end, Dr. Jay and his group have studied how the biophysical culture microenvironment of exosome-producing cells can be manipulated to control non-coding RNA cargo and bioactivity of exosomes. This investigation is potentially transformative in identifying critical parameters that could be controlled towards development of rationally designed, scalable biomanufacturing approaches for therapeutic exosomes. REU students will have the opportunity to employ both 3D printing and microsystems design to test specific hypotheses related to the roles of biophysical parameters on exosome production and bioactivity. REU participants would conduct the majority of their work at the University of Maryland.
(Dr. Weijie Chen, Food and Drug Administration)
As the cornerstone of modern medicine and cancer care, pathology is undergoing a revolutionary change from reviewing glass slides of tissue specimen under a microscope to displaying scanned digital whole-slide images (WSI) on monitors. The availability of large amount of WSI data and the rise of artificial intelligence (AI) and machine learning (ML) technologies have given rise to the field of AI in digital pathology, where AI/ML algorithms are developed for cancer detection, segmentation and quantification of cells or other structures of interest, and so on. The overall goal of this project is to develop assessment methodologies that enables collecting evidence of safety and effectiveness in a least burdensome fashion thereby facilitating the deployment of AI/ML technologies to pathology applications to improve diagnostic accuracy, efficiency, and patient management. We leverage the use of publicly available datasets and software code and develop image processing and AI/ML pipelines. We investigate study design and data analysis methods for characterizing the generalizability of AI/ML performance and its robustness to image acquisition variations due to e.g., scanner, lab, and site. The REU student will gain experiences in some of the following areas: data collection and management, programming, image processing and statistical analysis methodologies.
(Drs. Richard Gray & Pavel Takmakov, Food and Drug Administration)
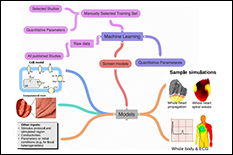 Sudden cardiac death resulting from ventricular fibrillation is the leading cause of death in the industrialized world. A better understanding of heart physiology, including individualized function, has the potential to prevent such deaths. Multi-scale models comprised of equations governing sub-cellular and cellular biochemical processes and bioelectric phenomena on the level of tissue, organ, and whole body are well understood and used to study life threatening cardiac arrhythmias including fibrillation. However, we have reached a “crises of complexity” because over a hundred cell models have been developed each with multitude of variables parameters. PubMed, a publication database, returns more than 140,000 articles for “cardiac models” as a keyword. Unfortunately, these models are highly overdetermined and rarely properly validated which prevents appropriate evaluation for a specific context of use that is necessary for clinical applications. We are in the process of automating the cardiac cell model development process to make it transparent, reproducible, and which avoids over-determined models. Cardiac cell models have always been ‘data-driven’ with thousands of experimental studies in different formats providing the basis for the models. In this project, we are using data science approaches to develop an algorithm to extract quantitative data from these experimental studies and use machine learning for building a framework to translate these data into robust cardiac cell models with a training set consisting of manually generated database of over a hundred of such studies. REU participants would conduct the majority of their work at the Food and Drug Administration.
Sudden cardiac death resulting from ventricular fibrillation is the leading cause of death in the industrialized world. A better understanding of heart physiology, including individualized function, has the potential to prevent such deaths. Multi-scale models comprised of equations governing sub-cellular and cellular biochemical processes and bioelectric phenomena on the level of tissue, organ, and whole body are well understood and used to study life threatening cardiac arrhythmias including fibrillation. However, we have reached a “crises of complexity” because over a hundred cell models have been developed each with multitude of variables parameters. PubMed, a publication database, returns more than 140,000 articles for “cardiac models” as a keyword. Unfortunately, these models are highly overdetermined and rarely properly validated which prevents appropriate evaluation for a specific context of use that is necessary for clinical applications. We are in the process of automating the cardiac cell model development process to make it transparent, reproducible, and which avoids over-determined models. Cardiac cell models have always been ‘data-driven’ with thousands of experimental studies in different formats providing the basis for the models. In this project, we are using data science approaches to develop an algorithm to extract quantitative data from these experimental studies and use machine learning for building a framework to translate these data into robust cardiac cell models with a training set consisting of manually generated database of over a hundred of such studies. REU participants would conduct the majority of their work at the Food and Drug Administration.
There are two projects in the Materials Characterization Laboratory.
Principal Investigator: Srilekha Das
In either case, the REU participant will learn how to measure viscoelastic properties of the subject material by dynamic mechanical analysis (DMA), along with necessary research management skills. Requirement: The trainee must be meticulous about his/her work style, maintaining lab-notebook and time management. [DMA knowledge is not required, instead, it will be taught during the training.]
Materials Characterization Project 1:
Viscoelastic properties of lung surface extra cellular matrix in the presence of surgical sealants used for preventing Pneumothoraces;
Materials Characterization Project 2:
Studying the durability of bioprosthetic heart valve in the presence of various cardiac enzymes.
(Dr. Shawn He, University of Maryland)
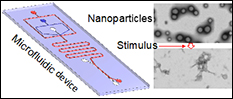 Stem cells are attracting ever-increasing attention for various biomedical applications including tissue engineering and regenerative medicine. However, the approaches used today for culturing stem cells in vitro are non-physiological and stem cells are banked in solutions containing highly toxic organic solvents (e.g., dimethylsulfoxide that is commonly known as DMSO). The former may induce genotypic and phenotypic changes in the stem cells, to compromise the quality of stem cells for tissue regeneration. The latter makes it indispensable to rigorously and tediously wash the stem cells after banking before implanting into patients, which may cause significant death or loss of the precious cells. The objective of this project is to develop 3D biomimetic culture of stem cells to minimize their spontaneous differentiation under in vitro culture, and further engineer the stem cells with micro and nanoscale biomaterials to achieve organic solvent-free banking so that they can be transplanted into patients without tedious washing. This will be achieved by utilizing the extensive nanotechnology approaches such as non-planar microfluidics and drug delivery with stimuli-responsive nanoparticles developed in the He Laboratory. REU students will learn how to fabricate microfluidic devices, use the devices to encapsulate stem cells for miniaturized 3D culture, use therapeutics-laden nanoparticles to engineer stem cells for banking, and make significant contributions to the innovative and impactful project. REU participants would conduct the majority of their work at the University of Maryland.
Stem cells are attracting ever-increasing attention for various biomedical applications including tissue engineering and regenerative medicine. However, the approaches used today for culturing stem cells in vitro are non-physiological and stem cells are banked in solutions containing highly toxic organic solvents (e.g., dimethylsulfoxide that is commonly known as DMSO). The former may induce genotypic and phenotypic changes in the stem cells, to compromise the quality of stem cells for tissue regeneration. The latter makes it indispensable to rigorously and tediously wash the stem cells after banking before implanting into patients, which may cause significant death or loss of the precious cells. The objective of this project is to develop 3D biomimetic culture of stem cells to minimize their spontaneous differentiation under in vitro culture, and further engineer the stem cells with micro and nanoscale biomaterials to achieve organic solvent-free banking so that they can be transplanted into patients without tedious washing. This will be achieved by utilizing the extensive nanotechnology approaches such as non-planar microfluidics and drug delivery with stimuli-responsive nanoparticles developed in the He Laboratory. REU students will learn how to fabricate microfluidic devices, use the devices to encapsulate stem cells for miniaturized 3D culture, use therapeutics-laden nanoparticles to engineer stem cells for banking, and make significant contributions to the innovative and impactful project. REU participants would conduct the majority of their work at the University of Maryland.
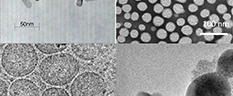
It is increasingly clear that rationally designed combination treatments impacting multiple targets will most likely improve outcomes in patients with advanced stage ovarian cancer. However, the selective delivery of multiple regimens to the right place, at the right time and in the correct sequence with consideration of mechanistic interaction remains a major challenge in combination treatment. Nanotechnology combined with light-activatable approaches provide a unique opportunity to deliver multiple agents that target several key molecular pathways. Dr. Huang’s team is working to develop a multi-compartment, light-activatable nanoplatform that can deliver three regimens in a unique manner—where one treatment primes the target for the second modality, and the subsequent evasion pathways are mitigated by a third agent; all agents are rationally selected and released in an appropriate time and sequence to account for mechanistic interactions and to improve outcomes. REU students participating in this project will have the opportunity to utilize Huang lab’s light-activatable nanoplatform to test new combination therapies in 3D ovarian cancer models. REU participants would conduct the majority of their work at the University of Maryland.
(Drs. Keaton Nahan, Berk Oktem, Samanthi Wickramasekara)
Volatile extractables in medical devices are a particular concern because they a) are most likely to migrate out of implanted materials, b) have greater potential toxicity due to their low molecular weight, and c) present an analytical challenge by being harder to identify and quantify using current chemical characterization screening approaches. Headspace GC/MS is a sample introduction and detection method for volatiles. This study aims to conduct solvent-less extraction of medical devices by headspace analysis, to develop a standardized method for identification and quantification of volatile extractables that would otherwise be lost to analysis if current methods were used. Student(s) involved in this project will conduct searches for relevant medical device literature, received hands-on training to perform headspace GC/MS, and learn how to conduct data analysis.
Vascular smooth muscle cells contract or relax to change blood vessel diameter, which is essential to healthy cardiovascular function. Recent studies have shown that the layer of fat surrounding most blood vessels, perivascular adipose tissue, may modulate vascular contractility. In healthy vessels, perivascular adipose tissue relaxes the adjacent blood vessel, making it easier for blood to flow. However, in diseases such hypertension and atherosclerosis, perivascular adipose tissue may cause the blood vessels to constrict. While blood vessel constriction and dilation can be measured in humans and animals, it is difficult to study the effects of perivascular adipose tissue on the blood vessels in these models.
The goal of this project is to create an artery-on-a-chip which includes perivascular adipose tissue and enables measurement of vascular contractility. Specifically, we are creating an artery within a hydrogel by circumferentially aligning contractile vascular smooth muscle cells around a hollow channel. We are then lining the channel with endothelial cells and surrounding the channel with perivascular adipose tissue. Once the device is built, we will thoroughly verify the device using biochemical assays and compare our results to mouse blood vessel measurements and to human studies. In the future, the artery-on-a-chip can be used for drug testing as well as to improve our understanding of how changes in perivascular adipose tissue affect vascular function.
REU students will learn how to create the microfluidic device, culture both endothelial and smooth muscle cells, and load them in the device. The students will also assess cell function under different mechanical and biochemical conditions. REU participants would conduct the majority of their research at the University of Maryland.
(Dr. Enusha Karunasena, Food and Drug Administration)
Wound infections are a reoccurring malignancy facing American healthcare: costs within the U.S. for wound-care can be upwards of $50 billion per year and improper wound care results in complications including wound debridement, surgical amputation or increased morbidity/mortality due to opportunistic infections (i.e. Staphylococcus aureus and Pseudomonas aeruginosa). For decades, the infection control laboratories within the Department of Biology, Chemistry, and Materials Science (DBCMS) have developed simulated fluids to mimic wounds and other biological test conditions to examine wound dressings. Building on this success: our goal is to address other pending concerns by measuring (1) whether antimicrobial properties remain intact for the duration of time described in product labeling (2) examine the direct effect of these dressings on adaptable traits expressed by bacteria that survive and (3) evaluate established molecular-markers for bacterial growth using real-time PCR (RT-PCR) assays to develop faster, quantitative, and reproducible methods to measure bacterial growth in wound dressing, for the FDA and its sponsors.
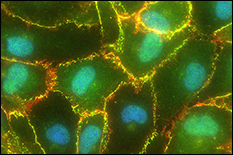 Tumor cell metastases to the brain are common in breast, skin, and lung cancer patients and are associated with high morbidity and poor prognosis. Blood-brain barrier (BBB) dysfunction is believed to occur in tumor metastasis, as well as infectious disease, stroke, multiple sclerosis, HIV, and Alzheimer’s disease. However, it is not understood how metastatic tumor cells are able to cross the BBB, given the BBB’s inherent function to tightly regulate flux of only select cellular and molecular materials into the brain’s interstitial fluid. A major hurdle preventing an understanding of the mechanisms of brain metastasis is the lack of physiologically-relevant BBB in vitro models. Dr. Stroka's team is working to develop a microsystem BBB-on-a-chip using microfabrication techniques and to use this model to understand brain endothelial cell (EC) biomechanics and the mechanobiology of brain metastasis. REU students participating in this project will have the opportunity to utilize the lab's BBB-microsystem to test hypotheses on the mechanisms of cancer metastasis. REU participants would conduct the majority of their work at the University of Maryland.
Tumor cell metastases to the brain are common in breast, skin, and lung cancer patients and are associated with high morbidity and poor prognosis. Blood-brain barrier (BBB) dysfunction is believed to occur in tumor metastasis, as well as infectious disease, stroke, multiple sclerosis, HIV, and Alzheimer’s disease. However, it is not understood how metastatic tumor cells are able to cross the BBB, given the BBB’s inherent function to tightly regulate flux of only select cellular and molecular materials into the brain’s interstitial fluid. A major hurdle preventing an understanding of the mechanisms of brain metastasis is the lack of physiologically-relevant BBB in vitro models. Dr. Stroka's team is working to develop a microsystem BBB-on-a-chip using microfabrication techniques and to use this model to understand brain endothelial cell (EC) biomechanics and the mechanobiology of brain metastasis. REU students participating in this project will have the opportunity to utilize the lab's BBB-microsystem to test hypotheses on the mechanisms of cancer metastasis. REU participants would conduct the majority of their work at the University of Maryland.
(Dr. Jeffrey L. Silberberg, Food and Drug Administration)
Physiological polysomnographic signals and data were recorded in a clinical setting in packed binary form from 10 neonatal subjects at the Nyquist sampling rate. The signals and data were recorded when the subjects were being evaluated for central apnea by way of overnight sleep studies. Technician annotations were also recorded. This project involves unpacking the data, performing a sin x / x resampling, scaling, time aligning the individual signals, and preparing and submitting the resulting waveforms and data to Physionet/Physiobank. Some processing software was written in HP Vee (now Keysight Vee); however, software such as LabView or MatLab could be used.
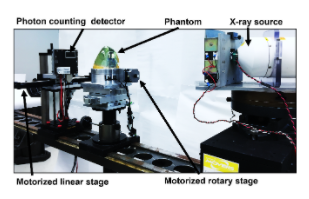
For over a century now, clinical x-ray imaging systems have operated in energy integrating mode. We are currently at the brink of a revolutionary change in clinical x-ray detector technology, where future detectors will be photon-counting, allowing for energy discrimination. These photon-counting detector systems will have numerous benefits especially in breast imaging because they provide a substantial increase in the raw imaging information that is available (e.g., multi-energy information). However, new algorithms are needed for using this information to increase the accuracy of diagnostic decisions. This proposed research project will focus on the investigation of new algorithms that can provide accurate a virtual breast biopsy (e.g., distinguishing malignant from benign lesions with spectral imaging). This internship will offer the opportunity to explore research and development career options, gain hands-on experience with radiation detection instrumentation and medical imaging physics. Students will have the opportunity to submit a poster in the annual FDA student poster session and co-author publications.
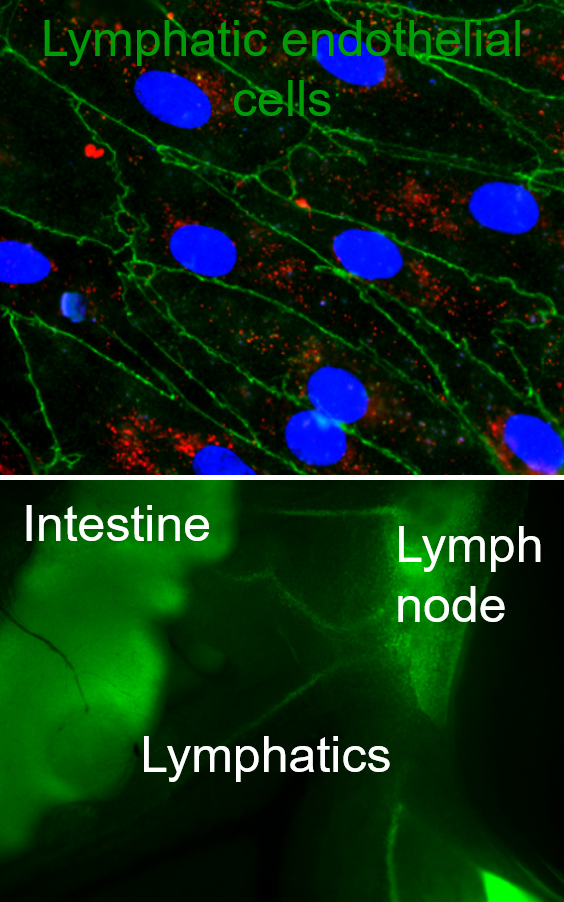 Lymphatics are the ‘sewer system’ and ‘highway for immune cells’ of the body, responsible for transporting fluids, solutes, and particulates away from the peripheral tissues via the lymph nodes into systemic circulation, ensuring the maintenance of tissue homeostasis and fluid balance between tissues and systemic circulation. Regulation of this fluid balance is crucial to ensure normal tissue functions and to regulate tissue functions in disease. Furthermore, in the past decade, lymphatics have been identified as a target for drug delivery, with lymphatic targeting nanoparticles enhancing immunotherapeutic and vaccine efficacies. However, we still do not know what beyond size governs lymphatic targeting, how targeting local lymphatic vessels that drain to specific lymph nodes can modulate immune responses, and additionally we do not have a clear understanding of the role that the stromal compartment plays in regulating of lymphatic transport in both homeostasis and disease. Our lab seeks to address these gaps in knowledge and develop new technologies such as in vitro model systems of mucosal surfaces, 3D lymphatic vessels, and drug delivery systems to target and study lymphatics, particularly focusing on lymphatic transport functions. REU students will learn to work with lymphatic endothelial cells, design complex gel and organoid culture as well as microfluidic systems, and loading cells into these systems. Additionally, students interested in drug delivery side of our work will explore designing nanocarriers with various surface chemistries, loading these carriers with drugs and biomacromolecules of interest, and studying how surface chemistry and targeting molecules can modulate particulate transport across epithelial and lymphatic layers. REU participants would conduct the majority of their research at the University of Maryland.
Lymphatics are the ‘sewer system’ and ‘highway for immune cells’ of the body, responsible for transporting fluids, solutes, and particulates away from the peripheral tissues via the lymph nodes into systemic circulation, ensuring the maintenance of tissue homeostasis and fluid balance between tissues and systemic circulation. Regulation of this fluid balance is crucial to ensure normal tissue functions and to regulate tissue functions in disease. Furthermore, in the past decade, lymphatics have been identified as a target for drug delivery, with lymphatic targeting nanoparticles enhancing immunotherapeutic and vaccine efficacies. However, we still do not know what beyond size governs lymphatic targeting, how targeting local lymphatic vessels that drain to specific lymph nodes can modulate immune responses, and additionally we do not have a clear understanding of the role that the stromal compartment plays in regulating of lymphatic transport in both homeostasis and disease. Our lab seeks to address these gaps in knowledge and develop new technologies such as in vitro model systems of mucosal surfaces, 3D lymphatic vessels, and drug delivery systems to target and study lymphatics, particularly focusing on lymphatic transport functions. REU students will learn to work with lymphatic endothelial cells, design complex gel and organoid culture as well as microfluidic systems, and loading cells into these systems. Additionally, students interested in drug delivery side of our work will explore designing nanocarriers with various surface chemistries, loading these carriers with drugs and biomacromolecules of interest, and studying how surface chemistry and targeting molecules can modulate particulate transport across epithelial and lymphatic layers. REU participants would conduct the majority of their research at the University of Maryland.
 The presence of amyloid beta plaques in the human brain is considered one of the hallmark features of Alzheimer's disease (AD) and a promising biomarker for early diagnosis. In early AD stages, Aβ plaques are formed when Aβ peptides aggregate and accumulate in the brain to form higher-order fibrils with β-sheet structures. A structurally sensitive and high-resolution imaging method is required to detect plaques in the human brain and allow timely therapeutic interventions to prevent AD progression. To this end, we are designing a novel amyloid imaging system prototype based on small-angle x-ray scattering (SAXS) to quantify amyloid load without contrast agents based on the nanometer-scale structural features of plaques. REU student will have an opportunity to prepare human brain phantoms with amyloid plaque models and assist in testing the feasibility of detecting amyloid plaques using a newly designed amyloid imaging spectral SAXS prototype.
The presence of amyloid beta plaques in the human brain is considered one of the hallmark features of Alzheimer's disease (AD) and a promising biomarker for early diagnosis. In early AD stages, Aβ plaques are formed when Aβ peptides aggregate and accumulate in the brain to form higher-order fibrils with β-sheet structures. A structurally sensitive and high-resolution imaging method is required to detect plaques in the human brain and allow timely therapeutic interventions to prevent AD progression. To this end, we are designing a novel amyloid imaging system prototype based on small-angle x-ray scattering (SAXS) to quantify amyloid load without contrast agents based on the nanometer-scale structural features of plaques. REU student will have an opportunity to prepare human brain phantoms with amyloid plaque models and assist in testing the feasibility of detecting amyloid plaques using a newly designed amyloid imaging spectral SAXS prototype. 
 The human gastrointestinal (GI) microbiome, often viewed as a complex organ itself, influences homeostasis and is implicated in many human diseases. There are few methodologies that enable resolution of its signaling, cell growth, and chemical environments even at the macro length scale. In the last several years however, researchers have developed micro- and meso-systems for interrogating GI tract biology. At the microscale, organ- or animal-on-a-chip methodologies have been developed for studying the GI tract. They provide first-of-their-kind access to biological function in user-controlled conditions. However, current approaches lack the ability to interrogate and modulate molecular signaling at cellular length scales and in real time. We have developed methodologies to combine synthetic biology with biofabrication to create smart hydrogels for interrogating molecular space. We do this by embedding “rewired” bacteria to serve as information translators into electrodeposited hydrogels that are assembled onto the sidewalls of a microfluidic channel (see Fig. 4). As a demonstration, we have assembled engineered E. coli to express blue fluorescent protein in response to a signal molecule, in this case the gratuitous lac operon inducer IPTG (Fig. 4c). In Fig. 4d, we show that one can layer cells using electrodeposition in configurations that enable visual access as well as chemical access so that cells can respond to molecular cues. In this REU project, using the same electrodeposition methodologies we have already developed in large systems, REU students will test the hypothesis that biologically active bacterial signaling molecules can be detected by a hydrogel bilayer that contains (i) signal sensing bacteria and (2) a catechol-modified chitosan layer incorporated onto the side wall of the channels shown in Fig. 4a,b. The students will assess whether the assembled cells can convert signal molecules to redox mediators in a manner that enables electrochemical detection. A successful result would be the first case of a biofabricated glucose sensor in an animal on a chip device. Such tests will be the first examples of the electrochemical interrogation of interkingdom biological signaling in an in vitro system. REU participants would conduct the majority of their work at the University of Maryland.
The human gastrointestinal (GI) microbiome, often viewed as a complex organ itself, influences homeostasis and is implicated in many human diseases. There are few methodologies that enable resolution of its signaling, cell growth, and chemical environments even at the macro length scale. In the last several years however, researchers have developed micro- and meso-systems for interrogating GI tract biology. At the microscale, organ- or animal-on-a-chip methodologies have been developed for studying the GI tract. They provide first-of-their-kind access to biological function in user-controlled conditions. However, current approaches lack the ability to interrogate and modulate molecular signaling at cellular length scales and in real time. We have developed methodologies to combine synthetic biology with biofabrication to create smart hydrogels for interrogating molecular space. We do this by embedding “rewired” bacteria to serve as information translators into electrodeposited hydrogels that are assembled onto the sidewalls of a microfluidic channel (see Fig. 4). As a demonstration, we have assembled engineered E. coli to express blue fluorescent protein in response to a signal molecule, in this case the gratuitous lac operon inducer IPTG (Fig. 4c). In Fig. 4d, we show that one can layer cells using electrodeposition in configurations that enable visual access as well as chemical access so that cells can respond to molecular cues. In this REU project, using the same electrodeposition methodologies we have already developed in large systems, REU students will test the hypothesis that biologically active bacterial signaling molecules can be detected by a hydrogel bilayer that contains (i) signal sensing bacteria and (2) a catechol-modified chitosan layer incorporated onto the side wall of the channels shown in Fig. 4a,b. The students will assess whether the assembled cells can convert signal molecules to redox mediators in a manner that enables electrochemical detection. A successful result would be the first case of a biofabricated glucose sensor in an animal on a chip device. Such tests will be the first examples of the electrochemical interrogation of interkingdom biological signaling in an in vitro system. REU participants would conduct the majority of their work at the University of Maryland. The constant generation and then movement of mucus from the lung airways to the throat, where it is then swallowed, is a key method by which the body protects against infection. However, in the airways of individuals with chronic lung diseases (e.g. asthma, chronic bronchitis, cystic fibrosis), build-up of mucus with abnormal biochemical and biophysical properties leads to airway obstruction and contributes to impaired lung function. Due to the natural heterogeneity of mucus derived from patients and animal models, we currently have a limited understanding of how mucus gel components impact the functional properties of mucus such as viscoelasticity for optimal mucus clearance and network architecture for pathogen/particulate capture. Toward this end, the goal of our NSF REU projects is to better understand the biophysical and biomolecular features of mucins that control their assembly into mucus. In our first REU student project, a bioengineered mucin-based hydrogel will be developed to mimic the physiological conditions under which the mucus gel network is assembled in health and disease. In a second project, an REU student will use CRISPR/Cas9 genetic engineering in order to design a tissue culture system with controlled airway mucus composition and function resembling that of a healthy and diseased airway. Together, this work will provide state-of-the art models that enable optimization and design of new therapies for chronic lung diseases. REU students engaged in this project will gain first-hand experience in biomaterial design, tissue culture, and/or genetic engineering. REU participants would conduct the majority of their work at the University of Maryland.
The constant generation and then movement of mucus from the lung airways to the throat, where it is then swallowed, is a key method by which the body protects against infection. However, in the airways of individuals with chronic lung diseases (e.g. asthma, chronic bronchitis, cystic fibrosis), build-up of mucus with abnormal biochemical and biophysical properties leads to airway obstruction and contributes to impaired lung function. Due to the natural heterogeneity of mucus derived from patients and animal models, we currently have a limited understanding of how mucus gel components impact the functional properties of mucus such as viscoelasticity for optimal mucus clearance and network architecture for pathogen/particulate capture. Toward this end, the goal of our NSF REU projects is to better understand the biophysical and biomolecular features of mucins that control their assembly into mucus. In our first REU student project, a bioengineered mucin-based hydrogel will be developed to mimic the physiological conditions under which the mucus gel network is assembled in health and disease. In a second project, an REU student will use CRISPR/Cas9 genetic engineering in order to design a tissue culture system with controlled airway mucus composition and function resembling that of a healthy and diseased airway. Together, this work will provide state-of-the art models that enable optimization and design of new therapies for chronic lung diseases. REU students engaged in this project will gain first-hand experience in biomaterial design, tissue culture, and/or genetic engineering. REU participants would conduct the majority of their work at the University of Maryland. Exosomes, a subset of cell-derived extracellular vesicles (EVs), have emerged as potential therapeutic vectors for a wide variety of applications. As alternatives to synthetic nanoparticles and cell therapies, exosomes have many potential benefits. Among these is the ability to transfer functional nucleic acids to recipient cells, as exosomes are physiological vectors for intercellular communication via nucleic acid transfer. However, exosomes also have shortcomings, especially their low potency due to containing relatively low amounts of nucleic acid cargo, especially non-coding RNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), which are critical to defining exosome bioactivity. Overcoming this limitation is crucial to enabling the therapeutic potential of exosomes. To this end, Dr. Jay and his group have studied how the biophysical culture microenvironment of exosome-producing cells can be manipulated to control non-coding RNA cargo and bioactivity of exosomes. This investigation is potentially transformative in identifying critical parameters that could be controlled towards development of rationally designed, scalable biomanufacturing approaches for therapeutic exosomes. REU students will have the opportunity to employ both 3D printing and microsystems design to test specific hypotheses related to the roles of biophysical parameters on exosome production and bioactivity. REU participants would conduct the majority of their work at the University of Maryland.
Exosomes, a subset of cell-derived extracellular vesicles (EVs), have emerged as potential therapeutic vectors for a wide variety of applications. As alternatives to synthetic nanoparticles and cell therapies, exosomes have many potential benefits. Among these is the ability to transfer functional nucleic acids to recipient cells, as exosomes are physiological vectors for intercellular communication via nucleic acid transfer. However, exosomes also have shortcomings, especially their low potency due to containing relatively low amounts of nucleic acid cargo, especially non-coding RNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), which are critical to defining exosome bioactivity. Overcoming this limitation is crucial to enabling the therapeutic potential of exosomes. To this end, Dr. Jay and his group have studied how the biophysical culture microenvironment of exosome-producing cells can be manipulated to control non-coding RNA cargo and bioactivity of exosomes. This investigation is potentially transformative in identifying critical parameters that could be controlled towards development of rationally designed, scalable biomanufacturing approaches for therapeutic exosomes. REU students will have the opportunity to employ both 3D printing and microsystems design to test specific hypotheses related to the roles of biophysical parameters on exosome production and bioactivity. REU participants would conduct the majority of their work at the University of Maryland. Sudden cardiac death resulting from ventricular fibrillation is the leading cause of death in the industrialized world. A better understanding of heart physiology, including individualized function, has the potential to prevent such deaths. Multi-scale models comprised of equations governing sub-cellular and cellular biochemical processes and bioelectric phenomena on the level of tissue, organ, and whole body are well understood and used to study life threatening cardiac arrhythmias including fibrillation. However, we have reached a “crises of complexity” because over a hundred cell models have been developed each with multitude of variables parameters. PubMed, a publication database, returns more than 140,000 articles for “cardiac models” as a keyword. Unfortunately, these models are highly overdetermined and rarely properly validated which prevents appropriate evaluation for a specific context of use that is necessary for clinical applications. We are in the process of automating the cardiac cell model development process to make it transparent, reproducible, and which avoids over-determined models. Cardiac cell models have always been ‘data-driven’ with thousands of experimental studies in different formats providing the basis for the models. In this project, we are using data science approaches to develop an algorithm to extract quantitative data from these experimental studies and use machine learning for building a framework to translate these data into robust cardiac cell models with a training set consisting of manually generated database of over a hundred of such studies. REU participants would conduct the majority of their work at the Food and Drug Administration.
Sudden cardiac death resulting from ventricular fibrillation is the leading cause of death in the industrialized world. A better understanding of heart physiology, including individualized function, has the potential to prevent such deaths. Multi-scale models comprised of equations governing sub-cellular and cellular biochemical processes and bioelectric phenomena on the level of tissue, organ, and whole body are well understood and used to study life threatening cardiac arrhythmias including fibrillation. However, we have reached a “crises of complexity” because over a hundred cell models have been developed each with multitude of variables parameters. PubMed, a publication database, returns more than 140,000 articles for “cardiac models” as a keyword. Unfortunately, these models are highly overdetermined and rarely properly validated which prevents appropriate evaluation for a specific context of use that is necessary for clinical applications. We are in the process of automating the cardiac cell model development process to make it transparent, reproducible, and which avoids over-determined models. Cardiac cell models have always been ‘data-driven’ with thousands of experimental studies in different formats providing the basis for the models. In this project, we are using data science approaches to develop an algorithm to extract quantitative data from these experimental studies and use machine learning for building a framework to translate these data into robust cardiac cell models with a training set consisting of manually generated database of over a hundred of such studies. REU participants would conduct the majority of their work at the Food and Drug Administration. Stem cells are attracting ever-increasing attention for various biomedical applications including tissue engineering and regenerative medicine. However, the approaches used today for culturing stem cells in vitro are non-physiological and stem cells are banked in solutions containing highly toxic organic solvents (e.g., dimethylsulfoxide that is commonly known as DMSO). The former may induce genotypic and phenotypic changes in the stem cells, to compromise the quality of stem cells for tissue regeneration. The latter makes it indispensable to rigorously and tediously wash the stem cells after banking before implanting into patients, which may cause significant death or loss of the precious cells. The objective of this project is to develop 3D biomimetic culture of stem cells to minimize their spontaneous differentiation under in vitro culture, and further engineer the stem cells with micro and nanoscale biomaterials to achieve organic solvent-free banking so that they can be transplanted into patients without tedious washing. This will be achieved by utilizing the extensive nanotechnology approaches such as non-planar microfluidics and drug delivery with stimuli-responsive nanoparticles developed in the He Laboratory. REU students will learn how to fabricate microfluidic devices, use the devices to encapsulate stem cells for miniaturized 3D culture, use therapeutics-laden nanoparticles to engineer stem cells for banking, and make significant contributions to the innovative and impactful project. REU participants would conduct the majority of their work at the University of Maryland.
Stem cells are attracting ever-increasing attention for various biomedical applications including tissue engineering and regenerative medicine. However, the approaches used today for culturing stem cells in vitro are non-physiological and stem cells are banked in solutions containing highly toxic organic solvents (e.g., dimethylsulfoxide that is commonly known as DMSO). The former may induce genotypic and phenotypic changes in the stem cells, to compromise the quality of stem cells for tissue regeneration. The latter makes it indispensable to rigorously and tediously wash the stem cells after banking before implanting into patients, which may cause significant death or loss of the precious cells. The objective of this project is to develop 3D biomimetic culture of stem cells to minimize their spontaneous differentiation under in vitro culture, and further engineer the stem cells with micro and nanoscale biomaterials to achieve organic solvent-free banking so that they can be transplanted into patients without tedious washing. This will be achieved by utilizing the extensive nanotechnology approaches such as non-planar microfluidics and drug delivery with stimuli-responsive nanoparticles developed in the He Laboratory. REU students will learn how to fabricate microfluidic devices, use the devices to encapsulate stem cells for miniaturized 3D culture, use therapeutics-laden nanoparticles to engineer stem cells for banking, and make significant contributions to the innovative and impactful project. REU participants would conduct the majority of their work at the University of Maryland.

 Tumor cell metastases to the brain are common in breast, skin, and lung cancer patients and are associated with high morbidity and poor prognosis. Blood-brain barrier (BBB) dysfunction is believed to occur in tumor metastasis, as well as infectious disease, stroke, multiple sclerosis, HIV, and Alzheimer’s disease. However, it is not understood how metastatic tumor cells are able to cross the BBB, given the BBB’s inherent function to tightly regulate flux of only select cellular and molecular materials into the brain’s interstitial fluid. A major hurdle preventing an understanding of the mechanisms of brain metastasis is the lack of physiologically-relevant BBB in vitro models. Dr. Stroka's team is working to develop a microsystem BBB-on-a-chip using microfabrication techniques and to use this model to understand brain endothelial cell (EC) biomechanics and the mechanobiology of brain metastasis. REU students participating in this project will have the opportunity to utilize the lab's BBB-microsystem to test hypotheses on the mechanisms of cancer metastasis. REU participants would conduct the majority of their work at the University of Maryland.
Tumor cell metastases to the brain are common in breast, skin, and lung cancer patients and are associated with high morbidity and poor prognosis. Blood-brain barrier (BBB) dysfunction is believed to occur in tumor metastasis, as well as infectious disease, stroke, multiple sclerosis, HIV, and Alzheimer’s disease. However, it is not understood how metastatic tumor cells are able to cross the BBB, given the BBB’s inherent function to tightly regulate flux of only select cellular and molecular materials into the brain’s interstitial fluid. A major hurdle preventing an understanding of the mechanisms of brain metastasis is the lack of physiologically-relevant BBB in vitro models. Dr. Stroka's team is working to develop a microsystem BBB-on-a-chip using microfabrication techniques and to use this model to understand brain endothelial cell (EC) biomechanics and the mechanobiology of brain metastasis. REU students participating in this project will have the opportunity to utilize the lab's BBB-microsystem to test hypotheses on the mechanisms of cancer metastasis. REU participants would conduct the majority of their work at the University of Maryland. For over a century now, clinical x-ray imaging systems have operated in energy integrating mode. We are currently at the brink of a revolutionary change in clinical x-ray detector technology, where future detectors will be photon-counting, allowing for energy discrimination. These photon-counting detector systems will have numerous benefits especially in breast imaging because they provide a substantial increase in the raw imaging information that is available (e.g., multi-energy information). However, new algorithms are needed for using this information to increase the accuracy of diagnostic decisions. This proposed research project will focus on the investigation of new algorithms that can provide accurate a virtual breast biopsy (e.g., distinguishing malignant from benign lesions with spectral imaging). This internship will offer the opportunity to explore research and development career options, gain hands-on experience with radiation detection instrumentation and medical imaging physics. Students will have the opportunity to submit a poster in the annual FDA student poster session and co-author publications.
For over a century now, clinical x-ray imaging systems have operated in energy integrating mode. We are currently at the brink of a revolutionary change in clinical x-ray detector technology, where future detectors will be photon-counting, allowing for energy discrimination. These photon-counting detector systems will have numerous benefits especially in breast imaging because they provide a substantial increase in the raw imaging information that is available (e.g., multi-energy information). However, new algorithms are needed for using this information to increase the accuracy of diagnostic decisions. This proposed research project will focus on the investigation of new algorithms that can provide accurate a virtual breast biopsy (e.g., distinguishing malignant from benign lesions with spectral imaging). This internship will offer the opportunity to explore research and development career options, gain hands-on experience with radiation detection instrumentation and medical imaging physics. Students will have the opportunity to submit a poster in the annual FDA student poster session and co-author publications. Lymphatics are the ‘sewer system’ and ‘highway for immune cells’ of the body, responsible for transporting fluids, solutes, and particulates away from the peripheral tissues via the lymph nodes into systemic circulation, ensuring the maintenance of tissue homeostasis and fluid balance between tissues and systemic circulation. Regulation of this fluid balance is crucial to ensure normal tissue functions and to regulate tissue functions in disease. Furthermore, in the past decade, lymphatics have been identified as a target for drug delivery, with lymphatic targeting nanoparticles enhancing immunotherapeutic and vaccine efficacies. However, we still do not know what beyond size governs lymphatic targeting, how targeting local lymphatic vessels that drain to specific lymph nodes can modulate immune responses, and additionally we do not have a clear understanding of the role that the stromal compartment plays in regulating of lymphatic transport in both homeostasis and disease. Our lab seeks to address these gaps in knowledge and develop new technologies such as in vitro model systems of mucosal surfaces, 3D lymphatic vessels, and drug delivery systems to target and study lymphatics, particularly focusing on lymphatic transport functions. REU students will learn to work with lymphatic endothelial cells, design complex gel and organoid culture as well as microfluidic systems, and loading cells into these systems. Additionally, students interested in drug delivery side of our work will explore designing nanocarriers with various surface chemistries, loading these carriers with drugs and biomacromolecules of interest, and studying how surface chemistry and targeting molecules can modulate particulate transport across epithelial and lymphatic layers. REU participants would conduct the majority of their research at the University of Maryland.
Lymphatics are the ‘sewer system’ and ‘highway for immune cells’ of the body, responsible for transporting fluids, solutes, and particulates away from the peripheral tissues via the lymph nodes into systemic circulation, ensuring the maintenance of tissue homeostasis and fluid balance between tissues and systemic circulation. Regulation of this fluid balance is crucial to ensure normal tissue functions and to regulate tissue functions in disease. Furthermore, in the past decade, lymphatics have been identified as a target for drug delivery, with lymphatic targeting nanoparticles enhancing immunotherapeutic and vaccine efficacies. However, we still do not know what beyond size governs lymphatic targeting, how targeting local lymphatic vessels that drain to specific lymph nodes can modulate immune responses, and additionally we do not have a clear understanding of the role that the stromal compartment plays in regulating of lymphatic transport in both homeostasis and disease. Our lab seeks to address these gaps in knowledge and develop new technologies such as in vitro model systems of mucosal surfaces, 3D lymphatic vessels, and drug delivery systems to target and study lymphatics, particularly focusing on lymphatic transport functions. REU students will learn to work with lymphatic endothelial cells, design complex gel and organoid culture as well as microfluidic systems, and loading cells into these systems. Additionally, students interested in drug delivery side of our work will explore designing nanocarriers with various surface chemistries, loading these carriers with drugs and biomacromolecules of interest, and studying how surface chemistry and targeting molecules can modulate particulate transport across epithelial and lymphatic layers. REU participants would conduct the majority of their research at the University of Maryland.